
Feingold, Mario
Growth and division of single bacterial cells; Time-lapse microscopy together with image processing; protein-DNA interaction at the single molecule level

Krichevsky, Oleg
Interactions and physical properties of biological molecules; Advanced optical and molecular biology techniques.

Meron, Ehud
Non linear dynamics and pattern formation; Complex systems; Modeling of dryland ecosystems; Ecosystem response to climate change; High-integrity human intervention …

Ashkenazy, Yosef
My research mainly deals with the physics of the climate system and the physics of sand dunes. I'm using both …

Beer, Avraham
Our lab combines a large variety of physical approaches, wet-lab biological based experiments, mathematical modeling, precise and innovative microscopy and …

Farago, Oded
My research combines analytical and computational methods of statistical mechanics to understand the physical principles behind the functioning of complex …

Golomb, David
I work in the fields of neurophysics and computational and theoretical neuroscience. I am interested in active sensing and dynamics …

Granek, Rony
I am a theoretical physicist and my research lies at the interface between soft matter physics and biophysics. I am …

Shamir, Maoz
Research field Computational neuroscience / Neurophysics Research Interests In my lab we apply tools and concepts from: Statistical Mechanics, Nonlinear …

Tsori, Yoav
Theory of soft matter physics on the mesoscopic scale (h-bar=0) - liquids, gels, colloid, membranes, polymers and liquid crystals. In …

Yochelis, Arik
Nonlinear physics of pattern forming systems: applications to energy devices, physical chemistry and biology

Sayag, Roiy
Dynamics of nonlinear fluids under weak friction, Instability theory, Geophysical flows, theoretical and experimental fluid dynamics.

Mordechai, Shaul
FTIR-Microspectroscopy; FTIR-spectroscopy;Fiber Evanescent Wave Spectroscopy
Research highlights
Single Cell Dynamics (Feingold's Group)
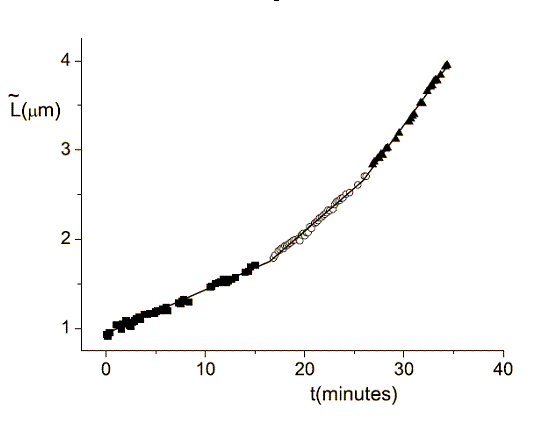
We use single cell phase-contrast and fluorescence time-lapse microscopy to monitor morphological changes during the division of E. coli. To bypass the limitations of optical resolution, we process the images using pixel intensity values for edge detection. We study the dynamics of the constriction width, W, and find that its formation starts shortly after birth much earlier than can be detected by simply viewing phase-contrast images. A simple geometrical model is shown to reproduce the behavior of W(t). Moreover, the time-dependence of the cell length, L(t), consists of three linear regimes.
Internal dynamics of biological polymers: DNA molecules, actin filaments (Krichevsky's Group)
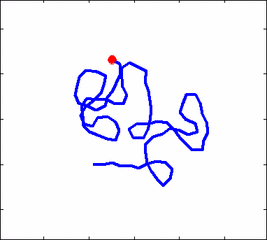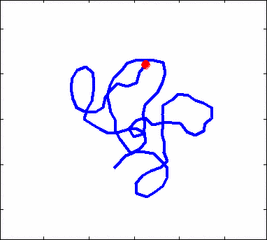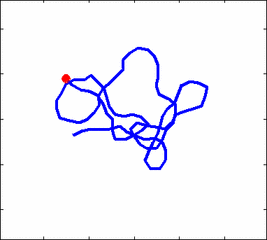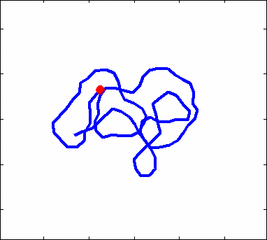
The problem of polymer dynamics is rather old, going back to the 1930-s.
How the stochastic thermal motion (diffusion) reveals itself in the dynamics of polymer segments which are bound by connectivity along the chain, by polymer stiffness, by topological constrains, by hydrodynamic and other interactions?
The question does not have simple solutions in neither theory, computer simulations, or experiments. We have developed an original experimental approach to measure the dynamics of biological polymers, such as DNA at the level of single monomer with high temporal and spatial resolution.
Mechanisms of Species Diversity Change in Stressed Environments (Meron's Group)
The impacts of environmental changes on species diversity, and thus on ecosystem function and stability, is a central topic of current ecological research. At the landscape scale, where symmetry breaking vegetation patterns appear, a transition from one pattern state to another may take place (Animation). Using a mathematical modeling approach, we developed a theory of plant communities in water limited system, and are currently using it to highlight mechanisms of species diversity change in response to climate changes and disturbances.
Nonlinear Dynamics and Biological Applications (Yochelis's Group)
Biological systems show a plethora of fascinating self-organized behaviors that range from organ to cellular levels, such as spiral waves, pulses, synchronization, and steady states that are periodic in space. These non-equilibrium phenomena emerge through either spontaneous or forced symmetry breaking mechanisms. Employing nonlinear dynamics methods, we attempt to understand specific cases (localized waves in the inner ear) as well as gain general insights into the emergence of traveling waves with motivation taken from molecular motors, actin polymerization and cardiac system.
Soft Matter Physics and Renewable Energy (Yochelis's Group)
Most renewable energy devices exploit nano-scale morphologies that are paramount to large surface area, required to increase activity. However, electrical effects are often strong enough to influence the structure of active layers of those materials leading to a notorious decrease in performance. To date, theoretical studies have dealt almost exclusively with uncoupled models of self-assembly and electrokinetics. We develop novel and computationally amenable mean-field frameworks that do unify them. Our expectations are to advance devices, such as batteries, supercapacitors, and solar cells.
Dynamics of Complex Fluids and Geophysical Flows (Sayag's Group)
The front of a fluid that displaces a less viscous fluid in shear-dominated flows is known to be stable. We show that in predominantly extensional flows on a sphere, a similar front of a strain-rate-softening fluid can become unstable and evolve fingerlike patterns comprised of rifts and tongues. The number of rifts and tongues declines with time and is selected by competition between interfacial hoop stress, geometric stretching, momentum dissipation, and spatial curvature. Our results elucidate fracture dynamics in complex fluids under extension and are applicable to a wide range of systems, including planetary-scale ice shelves as in snowball epochs and icy moons.